Exploring Novel Sources for Natural Products
The increasing speed at which – in particular multidrug resistant – pathogens spread poses a serious threat to the health and well-being of humanity. As a consequence new anti-infective drugs are urgently needed. Bacteria are prolific producers of low molecular weight compounds, so-called natural products, with antibiotic activity that can serve as basis for the development of novel drugs. However, traditional approaches to find antibiotics rarely lead to the discovery of truly novel structures as the rediscovery rate of already known compounds is very high.
1. Paleobiotechnology

Natural Products from Prehistoric Microbiomes
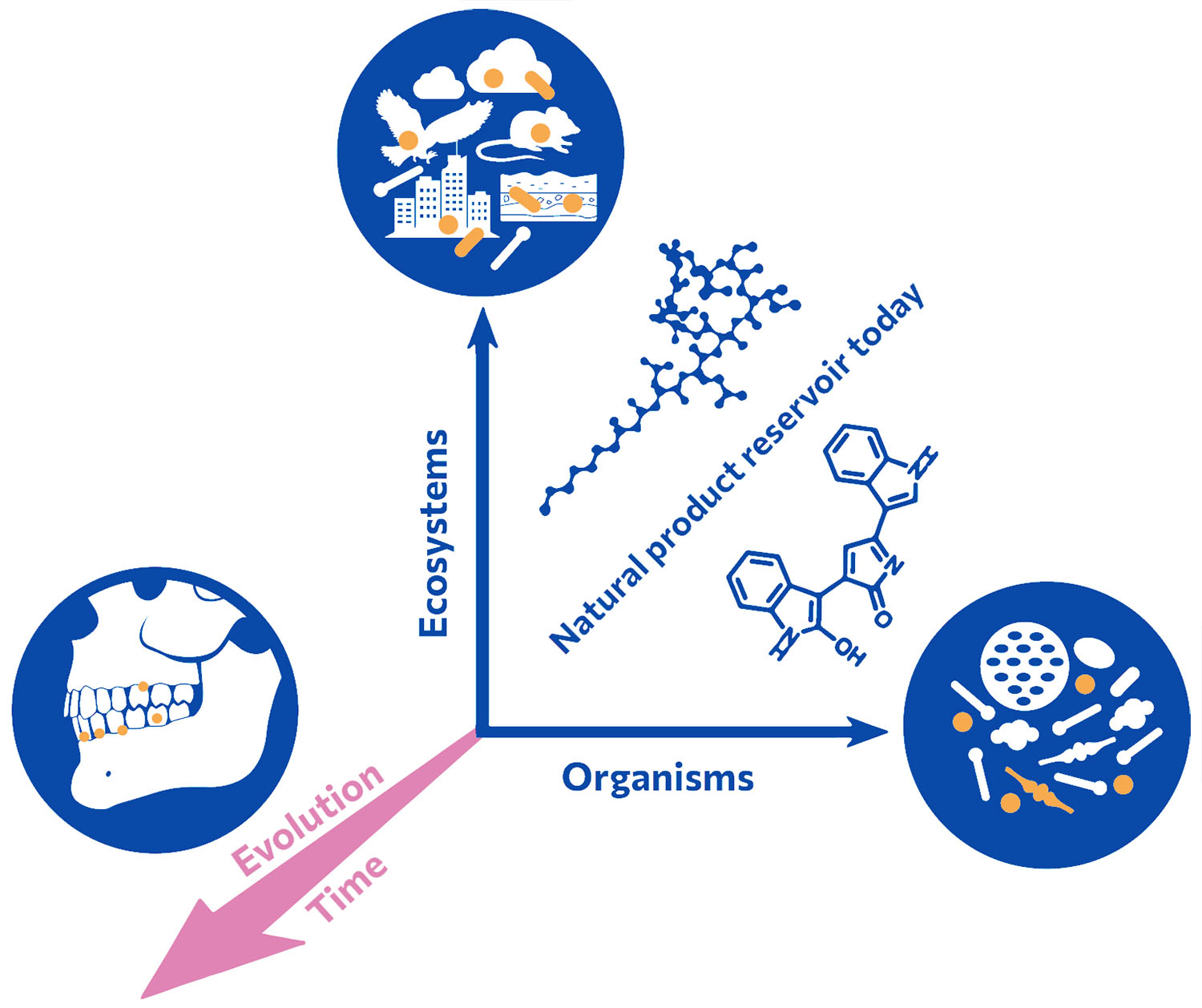
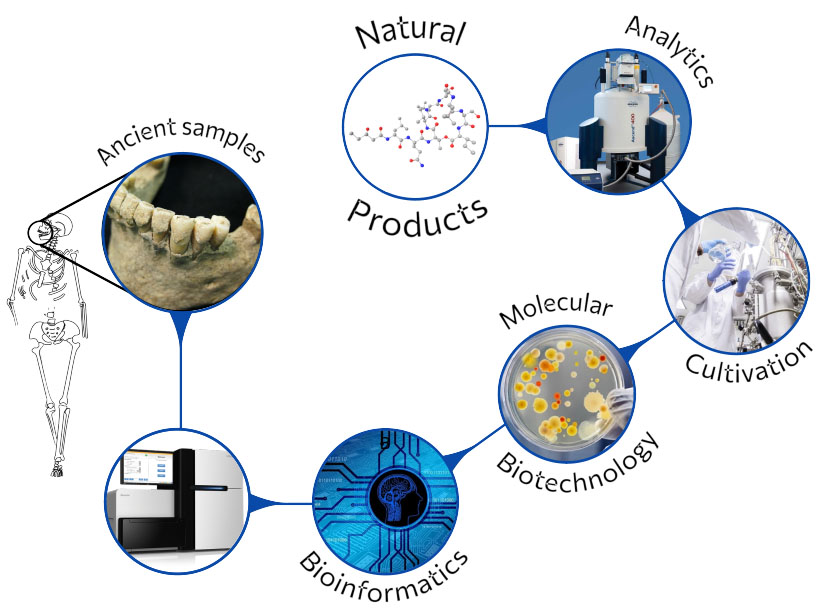
Our group accesses a so-far untapped source of natural product diversity. We do not limit our search for new molecules to the microorganisms that exist today, rather we investigate past microbiomes, effectively adding the time dimension to natural product discovery. We analyze the genetic information, i.e. ancient DNA, of prehistoric samples such as dental calculus or fecal remains. Metagenomes reflecting the past microbiome composition are assembled and ancient bacterial genomes are reconstructed. Within those we search for biosynthetic and other functional genes that we express in ‘modern’ microbial chassis. We also study the phylogeny and evolution of natural product biosynthetic genes.
In our research, we apply state-of-the art technologies for both (meta)genome assembly and analyses and for the identification and isolation of natural products. We also investigate traditionally produced food from regions of the world that have not been influenced by modern industry, which provide a promising starting point for identifying bioactive natural products. We closely collaborate with Prof. Dr. Christina Warinner from the Max Planck Institute for Evolutionary Anthropology in Leipzig / Harvard University, Cambridge, USA.
Antimicrobial Peptides from Ancient Microbiomes
Bacteriocins are a heterogeneous family of small, ribosomally synthesized, antimicrobial peptides produced by bacteria. They exert bacteriostatic or bacteriocidal activity mostly directed against bacteria in their surrounding habitat. The structural diversity of these peptides results in a broad spectrum of activities against bacteria, fungi, and viruses. The main industrial applications to date are as food preservatives. Additionally, they are also investigated as potential alternatives to classical antibiotics, especially because of their narrow antimicrobial activity spectrum. We extend the search for bacteriocins by investigating bacterial microbiomes from ancient sources and thereby add a temporal dimension. We analyze metagenomic data of genes coding for putative bacteriocins and subsequently the respective peptides are structurally and functionally analyzed using modern molecular biology methods.
Sampling the Past
Natural products and their biological functions are the result of millions of years of evolution. Their evolutionary diversification and intrinsic biological activities render them ideal candidates for medicinal lead structures. In addition to sampling diverse ecosystems and organisms for natural products and associated functions, we strive to travel back in time by analyzing ancient and preserved metagenomes from archaeological records and diverse cultures, respectively.
Combining Modern Technologies
Under certain circumstances, ancient DNA (aDNA) preserves well enough and reflects the microbial composition of the respective microbiome of the archaeological sample.
Next generation sequencing at low cost, as well as advances in the development of bioinformatics analyses makes it now feasible to link molecules or chemical moieties and resistance determinants to functional genes of ancient metagenomes. Gene synthesis and state-of-the-art cloning techniques using modern hosts finally allow to “resurrect” ancient functions.
2. Understanding Polymicrobial Associations
Characterizing Polymicrobial Interactions
Microorganisms are inherently gregarious organisms. In terrestrial habitats such as soil, they are in constant interaction with one another. Therefore, these complex habitats form a coliseum for microorganisms to train and gain functional abilities including secondary metabolite production. We want to understand and in particular visualize how bacteria interact both with each other as well as with eukaryotes such as amoebal predators or algae. We use in particular fluorescence in situ hybridization (FISH) as a powerful imaging technique to understand polymicrobial associations. Since FISH requires little to no genetic modifications of the organisms involved, it is perfectly suited to image microbes that are genetically not accessible. We combine different FISH variants with time-lapse imaging to identify morphological changes and variations in the population dynamics within complex associations. FISH is not restricted to prokaryotes; we use it to image other micro-eukaryotes for instance amoebae, algae, and fungi within both a pro- and eukaryotic background. In particular, we focus on:
- microbial predator-prey associations of the amoeba Dictyostelium discoideum and various bacteria of the genus Pseudomonas;
- Chlamydomonas–bacteria interactions;
- bacteria–fungi interactions.
Exploring the Microverse of the Plastisphere
Microplastic particles (small plastic debris less than 5 mm in size) have become a significant environmental concern due to their widespread presence in almost all habitats (oceans, soils, etc.). They serve as alternative substrates for biofilm formation that transport potential pathogens into novel habitats. Thus, they induce disbalance in various ecosystems while increasing the frequency of horizontal gene transfer and spreading of antibiotic resistance genes. Due to their small size, microplastic particles can be easily ingested by marine organisms, hence introducing pathogens to the food chain. For these reasons, it is crucial to establish an understanding of the implications of microplastic-associated microorganisms on the ecosystem and human health. However, many of the fundamental processes that underpin the formation and activities of plastic-colonizing biofilms remain poorly understood. We focus on investigating the interactions of commensal and pathogenic microorganisms with microplastic particles in the environment. We aim to understand:
- the factors driving microbial colonization on microplastic particles as well as the composition of resulting microbial biofilms;
- the role of extracellular communication molecules in microbial proliferation on microplastic; and
- the effects of biofilm formation on microplastic on horizontal gene transfer.
3. Natural Products in Amoebae–Bacteria Interactions
Investigating the metabolome associated with antagonistic ecological interactions between prokaryotes and eukaryotes is an effective approach to unlock the antimicrobial treasure trove of nature. Notably, amoeba–bacteria predator–prey interactions are fruitful sources of novel natural products. Amoeba are single-celled eukaryotes that prey on smaller bacteria. They are ubiquitous in soil and freshwater. As a consequence, bacteria sharing the same habitat as amoeba are exposed to a high selection pressure that fosters evolution of defense mechanisms such as the production of antimicrobial secondary metabolites.
In our group, we isolate bacteria from amoeba-rich environments and employ genome mining to detect biosynthetic pathways of new natural products using bioinformatics tools. In combination with molecular biology methods, we identify biosynthetic gene clusters that protect bacteria from amoebal grazing. We then isolate the corresponding natural products using state of the art fermentation techniques developed by the Leibniz-HKI Bio Pilot Plant Department and fully elucidate their structure through a combination of NMR, MS-MS, derivatization, and degradation experiments. In addition, we also use chemical synthesis to prepare analytical standards.
Finally, we examine the activity of the new natural products against a plethora of organisms (bacteria, fungi, amoeba, nematodes, mammalian cell lines) and explore the mode-of-action of potential lead natural products, which will aid the development of new anti-infectives.
4. Structure and Function of Polyketides in Dictyostelium discoideum
The genome of the social amoeba D. discoideum contains over 40 putative polyketide synthase (pks) genes. Their biosynthetic products as well as their functions are largely unknown. To identify and investigate these polyketides, we use a combination of molecular biology and analytical chemistry tools. Knockout mutants of selected pks genes are generated by modern genome editing technologies using CRISPR-Cas9 endonuclease and the resulting transcriptome and secondary metabolome are compared with the wildtype strain. Taken together with the documentation of phenotypic alterations of knockout mutants during their developmental process, the biological function of specific polyketides in D. discoideum can be predicted. Purification of specific polyketides from D. discoideum, which are often produced in small amounts in the amoeba, is achieved by overexpression of the respective PKS-coding genes. The chemical composition of the polyketides is elucidated and their biological activities are tested.