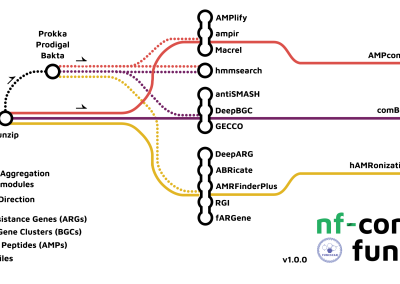
Big day for the developers of the bioinformatics pipeline in the Stallforth lab: The nextflow pipeline nf-core/funcscan – developed since late 2021 – got its first tagged release!
The pipeline will enable us (and others) to efficiently and reproducibly identify antimicrobial peptide sequences, antibiotic resistance genes and biosynthetic gene clusters in metagenomic sequences of bacterial samples. Watch a short pipeline introduction on YouTube (in the framework of the nf-core/bytesize lectures).
nf-core/funcscan v1.0.0 is now available under DOI 10.5281/zenodo.7643099 (see also the project ![]() repository).
repository).
The development team was going through the final steps of the first release together (and finally pressing the “Publish release” button) via gather.town. After successful pre- and post-release steps we celebrated with self-made blueberry cake!